Chapter 13 Data Wrangling with tidyverse
13.1 Welcome to the tidyverse

Figure 13.1: Artwork by Allison Horst
The tidyverse is a collection of R packages designed to facilitate data science. The so-called core tidyverse includes the following packages:
dplyrfor data manipulation;tidyrfor tidying data;ggplot2for plotting;readrfor reading in data files of various formats;stringrfor manipulating character strings;tibblefor a re-engineered alternative to data frames;purrrfor functional programming;forcatsfor better handling categorical variables.
While these are distinct packages and can be installed and loaded separately,
they share common grammar, syntax, and data structures. Most of the functions we
are going to see in this and the following Chapter are from dplyr and
ggplot2, but we’ll sprinkle some tidyr, stringr, and tibble here and
there. On top of the core packages, there are several other non-core
packages that are installed together with the tidyverse but are not
automatically loaded. These package are specialized on a few niche roles that
users may not always need.
To install the tidyverse, go ahead and run the following line:
Then load the package using:
## ── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
## ✔ dplyr 1.1.4 ✔ readr 2.1.4
## ✔ forcats 1.0.0 ✔ stringr 1.5.1
## ✔ ggplot2 3.4.4 ✔ tibble 3.2.1
## ✔ lubridate 1.9.3 ✔ tidyr 1.3.0
## ✔ purrr 1.0.2
## ── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
## ✖ dplyr::filter() masks stats::filter()
## ✖ dplyr::lag() masks stats::lag()
## ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errors13.2 Tidyverse functions
To demonstrate the use of tidyverse functions, we are going to work on the dragons dataset we used in Chapters 5, 6, and 7. Let’s load our data in using RSQLite like we learned in Chapter 7:
And let’s load all the tables:
dragons <- dbGetQuery(dragons_db, "SELECT * FROM dragons;")
capture_sites <- dbGetQuery(dragons_db, "SELECT * FROM capture_sites;")
captures <- dbGetQuery(dragons_db, "SELECT * FROM captures;")
morphometrics <- dbGetQuery(dragons_db, "SELECT * FROM morphometrics;")
diet <- dbGetQuery(dragons_db, "SELECT * FROM diet;")
tags <- dbGetQuery(dragons_db, "SELECT * FROM tags;")
deployments <- dbGetQuery(dragons_db, "SELECT * FROM deployments;")
telemetry <- dbGetQuery(dragons_db, "SELECT * FROM gps_data;")13.2.1 Subsetting columns
Let’s start practicing on the morphometrics table.
## [1] "data.frame"## measurement_id dragon_id date total_body_length_cm wingspan_cm
## 1 1 D96 2012-10-10 1069.8965 1389.5527
## 2 2 D400 2003-07-03 333.4600 634.9109
## 3 3 D316 2017-09-08 866.8935 1052.3702
## 4 4 D317 2016-09-05 1146.9708 1356.8084
## 5 5 D484 2016-12-04 1032.0520 1720.8641
## 6 6 D149 2012-02-13 919.9908 1533.5991
## tail_length_cm tarsus_length_cm claw_length_cm
## 1 595.2706 121.65175 15.596219
## 2 104.2241 38.10844 4.305086
## 3 373.7619 68.16869 12.719697
## 4 542.5670 172.43663 14.809363
## 5 596.4419 114.05057 11.985672
## 6 563.9201 134.18051 11.005070We can subset columns of interest from a table using the select function. The
function takes as arguments 1. the data and 2. the name/s of one or more
columns that we want to keep:
body_wing <- select(morphometrics, dragon_id, date, total_body_length_cm, wingspan_cm)
head(body_wing)## dragon_id date total_body_length_cm wingspan_cm
## 1 D96 2012-10-10 1069.8965 1389.5527
## 2 D400 2003-07-03 333.4600 634.9109
## 3 D316 2017-09-08 866.8935 1052.3702
## 4 D317 2016-09-05 1146.9708 1356.8084
## 5 D484 2016-12-04 1032.0520 1720.8641
## 6 D149 2012-02-13 919.9908 1533.5991The measurement_id column is the primary key of the morphometrics table. This
was important to have in the database but we may not want to keep that column
now that we are processing the data in R. We could drop that column by
selecting all the other ones, but that would be a lot of typing. There is a more
convenient way: we can use select to discard columns by adding a - in front
of their name:
## dragon_id date total_body_length_cm wingspan_cm tail_length_cm
## 1 D96 2012-10-10 1069.8965 1389.5527 595.2706
## 2 D400 2003-07-03 333.4600 634.9109 104.2241
## 3 D316 2017-09-08 866.8935 1052.3702 373.7619
## 4 D317 2016-09-05 1146.9708 1356.8084 542.5670
## 5 D484 2016-12-04 1032.0520 1720.8641 596.4419
## 6 D149 2012-02-13 919.9908 1533.5991 563.9201
## tarsus_length_cm claw_length_cm
## 1 121.65175 15.596219
## 2 38.10844 4.305086
## 3 68.16869 12.719697
## 4 172.43663 14.809363
## 5 114.05057 11.985672
## 6 134.18051 11.00507013.2.2 Subsetting rows with logical conditions
To subset rows of a table based on certain conditions, we can use the filter
function. For example, to filter only wingspans larger than 10 m:
## measurement_id dragon_id date total_body_length_cm wingspan_cm
## 1 1 D96 2012-10-10 1069.8965 1389.553
## 2 3 D316 2017-09-08 866.8935 1052.370
## 3 4 D317 2016-09-05 1146.9708 1356.808
## 4 5 D484 2016-12-04 1032.0520 1720.864
## 5 6 D149 2012-02-13 919.9908 1533.599
## 6 9 D283 2007-06-21 1698.1918 1387.194
## tail_length_cm tarsus_length_cm claw_length_cm
## 1 595.2706 121.65175 15.59622
## 2 373.7619 68.16869 12.71970
## 3 542.5670 172.43663 14.80936
## 4 596.4419 114.05057 11.98567
## 5 563.9201 134.18051 11.00507
## 6 666.4246 147.44219 13.1892313.2.3 Concatenating operations with pipes
One of the most revolutionary innovations that the tidyverse brings to R is this
operator: %>%, the pipe. A pipe is a connector that allows you to concatenate
subsequent actions into one single chunk of code. For instance, if we wanted to
drop the measurement_id column from the morphometrics table and filter
rows where wingspan_cm is larger than 10 m, this is what we would do without
the pipe:
no_pkey <- select(morphometrics, -measurement_id)
larger_than_10m <- filter(no_pkey, wingspan_cm > 1000)Instead, we can concatenate the two actions:
larger_than_10m <- morphometrics %>%
select(-measurement_id) %>%
filter(wingspan_cm > 1000)
head(larger_than_10m) ## dragon_id date total_body_length_cm wingspan_cm tail_length_cm
## 1 D96 2012-10-10 1069.8965 1389.553 595.2706
## 2 D316 2017-09-08 866.8935 1052.370 373.7619
## 3 D317 2016-09-05 1146.9708 1356.808 542.5670
## 4 D484 2016-12-04 1032.0520 1720.864 596.4419
## 5 D149 2012-02-13 919.9908 1533.599 563.9201
## 6 D283 2007-06-21 1698.1918 1387.194 666.4246
## tarsus_length_cm claw_length_cm
## 1 121.65175 15.59622
## 2 68.16869 12.71970
## 3 172.43663 14.80936
## 4 114.05057 11.98567
## 5 134.18051 11.00507
## 6 147.44219 13.18923The pipe takes the output of the previous line and feeds it as input into the
next one. Notice that you don’t have to repeat the name of the data object,
because the data is whatever the pipe is feeding into the function. There are
several advantages to using pipes compared to traditional syntax. First, by
using a pipe in the example above, we avoided saving intermediate objects (e.g.,
no_pkey) to the environment: we only saved the final result we wanted. Second,
we typed less. Third, our code is more readable because the syntax of our code
reflects the logical structure of what we are doing. You can read the pipe as
then: take the morphometrics table, then drop the measurement ID, then
filter records with wingspan larger than 10 m. The shortcut for inserting a pipe
is Ctrl + Shift + M on Windows and Cmd + Shift + M on Mac.
13.2.4 Creating new columns

Figure 13.2: Artwork by Allison Horst
The filtering we did above was pretty inconvenient because we had to calculate
the conversion between meters and centimeters in our head before applying the
wingspan filter. It would be easier to convert the column to meters to begin
with. We can create a new column using mutate:
larger_than_10m <- morphometrics %>%
select(-measurement_id) %>%
mutate(wingspan_m = wingspan_cm/100) %>%
filter(wingspan_m > 10)
head(larger_than_10m)## dragon_id date total_body_length_cm wingspan_cm tail_length_cm
## 1 D96 2012-10-10 1069.8965 1389.553 595.2706
## 2 D316 2017-09-08 866.8935 1052.370 373.7619
## 3 D317 2016-09-05 1146.9708 1356.808 542.5670
## 4 D484 2016-12-04 1032.0520 1720.864 596.4419
## 5 D149 2012-02-13 919.9908 1533.599 563.9201
## 6 D283 2007-06-21 1698.1918 1387.194 666.4246
## tarsus_length_cm claw_length_cm wingspan_m
## 1 121.65175 15.59622 13.89553
## 2 68.16869 12.71970 10.52370
## 3 172.43663 14.80936 13.56808
## 4 114.05057 11.98567 17.20864
## 5 134.18051 11.00507 15.33599
## 6 147.44219 13.18923 13.8719413.2.5 Limiting results
I have been saving each of my queries as an object and then doing head() to
show the first 6 rows of the result. Instead, I can save myself some typing and
space in my environment (and make everything look cleaner) by adding another
pipe at the end with the slice function. The slice function selects rows
based on position, so if I want to look at the first 6 I can do:
morphometrics %>%
select(-measurement_id) %>%
mutate(wingspan_m = wingspan_cm/100) %>%
filter(wingspan_m > 10) %>%
slice(1:6)## dragon_id date total_body_length_cm wingspan_cm tail_length_cm
## 1 D96 2012-10-10 1069.8965 1389.553 595.2706
## 2 D316 2017-09-08 866.8935 1052.370 373.7619
## 3 D317 2016-09-05 1146.9708 1356.808 542.5670
## 4 D484 2016-12-04 1032.0520 1720.864 596.4419
## 5 D149 2012-02-13 919.9908 1533.599 563.9201
## 6 D283 2007-06-21 1698.1918 1387.194 666.4246
## tarsus_length_cm claw_length_cm wingspan_m
## 1 121.65175 15.59622 13.89553
## 2 68.16869 12.71970 10.52370
## 3 172.43663 14.80936 13.56808
## 4 114.05057 11.98567 17.20864
## 5 134.18051 11.00507 15.33599
## 6 147.44219 13.18923 13.87194More generally, slice can be used as the equivalent of selecting rows from a
data frame using indexes like we did in Chapter 10. If I want
row 38 of the morphometrics table, I can do:
## measurement_id dragon_id date total_body_length_cm wingspan_cm
## 1 38 D367 2006-06-12 1129.259 1315.82
## tail_length_cm tarsus_length_cm claw_length_cm
## 1 578.4428 133.3162 24.20511This works with multiple rows too, and they don’t need to be consecutive:
## measurement_id dragon_id date total_body_length_cm wingspan_cm
## 1 38 D367 2006-06-12 1129.2590 1315.8203
## 2 12 D322 2009-03-09 309.3959 595.7932
## 3 84 D351 2015-10-04 1439.1528 1213.9245
## tail_length_cm tarsus_length_cm claw_length_cm
## 1 578.4428 133.31621 24.205108
## 2 137.9718 40.53882 4.273016
## 3 610.6215 110.42981 12.50282213.2.6 Tibbles
Another way to limit the output that gets printed to the console is to use
tibbles instead of data frames. A tibble is tidyverse’s data structure for
tabular data, so it is the tidyverse equivalent of a data frame. Tibbles
implement some functionalities that make working with them a bit more foolproof
than working with data frames (e.g., they’ll return an error instead of NULL
if you try to access a column that does not exist), but 99% of the time you
won’t notice the difference. This is because all tidyverse functions (as well as
some base R ones) work just the same with data frames and tibbles. However,
tibbles do have an advantage: when printing a tibble to the console, only the
first 10 records will show up, which is convenient because it means you don’t
need to use head() to prevent your console from being flooded with output.
Case in point:
morphometrics %>%
as_tibble() %>%
select(-measurement_id) %>%
mutate(wingspan_m = wingspan_cm/100) %>%
filter(wingspan_m > 10) ## # A tibble: 192 × 8
## dragon_id date total_body_length_cm wingspan_cm tail_length_cm
## <chr> <chr> <dbl> <dbl> <dbl>
## 1 D96 2012-10-10 1070. 1390. 595.
## 2 D316 2017-09-08 867. 1052. 374.
## 3 D317 2016-09-05 1147. 1357. 543.
## 4 D484 2016-12-04 1032. 1721. 596.
## 5 D149 2012-02-13 920. 1534. 564.
## 6 D283 2007-06-21 1698. 1387. 666.
## 7 D485 2002-10-04 957. 1780. 561.
## 8 D343 2016-04-08 1521. 1538. 618.
## 9 D237 2009-02-22 1205. 2120. 656.
## 10 D312 2001-06-16 927. 1268. 453.
## # ℹ 182 more rows
## # ℹ 3 more variables: tarsus_length_cm <dbl>, claw_length_cm <dbl>,
## # wingspan_m <dbl>As you can see, the appearance of a tibble is very similar to the one of a data frame. In fact, objects can be of multiple classes in R, and tibbles are also data frames under the hood!
## [1] "tbl_df" "tbl" "data.frame"Converting between tibble and data frame is also super easy, so you can switch back and forth between them whenever you need:
This last bit is useful to know especially when you are using other packages that were not written using tidyverse and strictly require data frames as input.
13.2.7 Joining tables
In Chapter 6, we talked about SQL joins. The concept of a join is not
exclusive to SQL, and in fact the tidyverse has functions that serve the same
exact purpose of SQL joins. Let’s get familiar with the one you’ll use most
often, left_join. As a reminder, a left join keeps all the rows of the left
table (the one you mention first) while attaching information from the right
table (the second one) whenever available. If no information is available, the
columns coming from the right table will have NA but the row will be retained.
To join two tables, these need to share at least one column. Let’s join the
morphometrics table with the dragons table:
## # A tibble: 327 × 12
## measurement_id dragon_id date total_body_length_cm wingspan_cm
## <int> <chr> <chr> <dbl> <dbl>
## 1 1 D96 2012-10-10 1070. 1390.
## 2 2 D400 2003-07-03 333. 635.
## 3 3 D316 2017-09-08 867. 1052.
## 4 4 D317 2016-09-05 1147. 1357.
## 5 5 D484 2016-12-04 1032. 1721.
## 6 6 D149 2012-02-13 920. 1534.
## 7 7 D285 2016-03-23 305. 699.
## 8 8 D256 2013-09-07 359. 652.
## 9 9 D283 2007-06-21 1698. 1387.
## 10 10 D213 2001-12-12 354. 671.
## # ℹ 317 more rows
## # ℹ 7 more variables: tail_length_cm <dbl>, tarsus_length_cm <dbl>,
## # claw_length_cm <dbl>, sex <chr>, age_class <chr>, species <chr>,
## # update_timestamp <chr>In this case the shared column has the same name in the two tables. If it doesn’t, you can still join based on that column. Let’s demonstrate how:
morphometrics %>%
as_tibble() %>%
rename(dragon = dragon_id) %>% # now the dragon_id column is called "dragon"
left_join(dragons, by = c("dragon" = "dragon_id")) ## # A tibble: 327 × 12
## measurement_id dragon date total_body_length_cm wingspan_cm tail_length_cm
## <int> <chr> <chr> <dbl> <dbl> <dbl>
## 1 1 D96 2012-1… 1070. 1390. 595.
## 2 2 D400 2003-0… 333. 635. 104.
## 3 3 D316 2017-0… 867. 1052. 374.
## 4 4 D317 2016-0… 1147. 1357. 543.
## 5 5 D484 2016-1… 1032. 1721. 596.
## 6 6 D149 2012-0… 920. 1534. 564.
## 7 7 D285 2016-0… 305. 699. 116.
## 8 8 D256 2013-0… 359. 652. 148.
## 9 9 D283 2007-0… 1698. 1387. 666.
## 10 10 D213 2001-1… 354. 671. 140.
## # ℹ 317 more rows
## # ℹ 6 more variables: tarsus_length_cm <dbl>, claw_length_cm <dbl>, sex <chr>,
## # age_class <chr>, species <chr>, update_timestamp <chr>So the syntax in the by argument when the names are different is
column_name_in_left_table = column_name_in_right_table.
13.2.8 Changing the order of columns
Incidentally, I have just shown how to change the name of a column by using
rename. We can also change the order in which columns appear in our table
using relocate. For example, say that we want to move the age class column
to right after the dragon ID:
morphometrics %>%
as_tibble() %>%
left_join(dragons, by = "dragon_id") %>%
relocate(age_class, .after = dragon_id)## # A tibble: 327 × 12
## measurement_id dragon_id age_class date total_body_length_cm wingspan_cm
## <int> <chr> <chr> <chr> <dbl> <dbl>
## 1 1 D96 Adult 2012-10-… 1070. 1390.
## 2 2 D400 Juvenile 2003-07-… 333. 635.
## 3 3 D316 Subadult 2017-09-… 867. 1052.
## 4 4 D317 Adult 2016-09-… 1147. 1357.
## 5 5 D484 Adult 2016-12-… 1032. 1721.
## 6 6 D149 Adult 2012-02-… 920. 1534.
## 7 7 D285 Juvenile 2016-03-… 305. 699.
## 8 8 D256 Juvenile 2013-09-… 359. 652.
## 9 9 D283 Adult 2007-06-… 1698. 1387.
## 10 10 D213 Juvenile 2001-12-… 354. 671.
## # ℹ 317 more rows
## # ℹ 6 more variables: tail_length_cm <dbl>, tarsus_length_cm <dbl>,
## # claw_length_cm <dbl>, sex <chr>, species <chr>, update_timestamp <chr>13.2.9 Calculations by group
Now that we know how to join the morphometrics table to the dragons table we can
calculate some summary statistics based on different groups. To do so, we use
the function group_by together with the function summarize. For instance,
let’s calculate minimum, maximum, and mean tail length for dragons of different
age classes:
morphometrics %>%
as_tibble() %>%
left_join(dragons, by = "dragon_id") %>%
group_by(age_class) %>%
summarize(min_tail_length_cm = min(tail_length_cm),
mean_tail_length_cm = mean(tail_length_cm),
max_tail_length_cm = max(tail_length_cm))## # A tibble: 3 × 4
## age_class min_tail_length_cm mean_tail_length_cm max_tail_length_cm
## <chr> <dbl> <dbl> <dbl>
## 1 Adult 502. 600. 686.
## 2 Juvenile 84.0 119. 148.
## 3 Subadult 338. 435. 544.We can also use group_by to count how many records we have for each group.
Let’s say we want to know how many dragons of each species we captured:
morphometrics %>%
as_tibble() %>%
left_join(dragons, by = "dragon_id") %>%
group_by(species) %>%
tally()## # A tibble: 10 × 2
## species n
## <chr> <int>
## 1 Antipodean Opaleye 16
## 2 Chinese Fireball 16
## 3 Common Welsh Green 51
## 4 Hebridean Black 42
## 5 Hungarian Horntail 29
## 6 Norwegian Ridgeback 59
## 7 Peruvian Vipertooth 39
## 8 Romanian Longhorn 44
## 9 Swedish Short-Snout 14
## 10 Ukrainian Ironbelly 1713.2.10 Sorting results
Let’s sort results of our count by species in decreasing order:
morphometrics %>%
as_tibble() %>%
left_join(dragons, by = "dragon_id") %>%
group_by(species) %>%
tally() %>%
arrange(n)## # A tibble: 10 × 2
## species n
## <chr> <int>
## 1 Swedish Short-Snout 14
## 2 Antipodean Opaleye 16
## 3 Chinese Fireball 16
## 4 Ukrainian Ironbelly 17
## 5 Hungarian Horntail 29
## 6 Peruvian Vipertooth 39
## 7 Hebridean Black 42
## 8 Romanian Longhorn 44
## 9 Common Welsh Green 51
## 10 Norwegian Ridgeback 59Nope, that didn’t do it. The species are sorted from the least to the most numerous (the default). We need to specify that we want to sort results in descending order:
morphometrics %>%
as_tibble() %>%
left_join(dragons, by = "dragon_id") %>%
group_by(species) %>%
tally() %>%
arrange(desc(n))## # A tibble: 10 × 2
## species n
## <chr> <int>
## 1 Norwegian Ridgeback 59
## 2 Common Welsh Green 51
## 3 Romanian Longhorn 44
## 4 Hebridean Black 42
## 5 Peruvian Vipertooth 39
## 6 Hungarian Horntail 29
## 7 Ukrainian Ironbelly 17
## 8 Antipodean Opaleye 16
## 9 Chinese Fireball 16
## 10 Swedish Short-Snout 1413.2.11 Extracting columns as vectors
Now say that we want to only keep morphometric measurements for dragons of species for which we have at least 30 individuals. We need to filter those whose species falls within that group. So this task can be divided in two steps: first, identify the group and store the result; second, filter the table based on records from those groups. Let’s see:
(species_over30 <- dragons %>%
group_by(species) %>%
tally() %>%
arrange(desc(n)) %>%
filter(n > 30) %>%
pull(species))## [1] "Norwegian Ridgeback" "Common Welsh Green" "Romanian Longhorn"
## [4] "Hebridean Black" "Peruvian Vipertooth" "Hungarian Horntail"The function pull at the end extracts the values in my column of interest and
returns them as a vector.
## [1] "character"If I used select instead, the result would be a tibble:
dragons %>%
group_by(species) %>%
tally() %>%
arrange(desc(n)) %>%
filter(n > 30) %>%
select(species)## # A tibble: 6 × 1
## species
## <chr>
## 1 Norwegian Ridgeback
## 2 Common Welsh Green
## 3 Romanian Longhorn
## 4 Hebridean Black
## 5 Peruvian Vipertooth
## 6 Hungarian Horntaildragons %>%
group_by(species) %>%
tally() %>%
arrange(desc(n)) %>%
filter(n > 30) %>%
select(species) %>%
class()## [1] "tbl_df" "tbl" "data.frame"Now that we have a vector with the species that we want to retain, we can apply our filter:
morphometrics %>%
as_tibble() %>%
left_join(dragons, by = "dragon_id") %>%
filter(species %in% species_over30)## # A tibble: 264 × 12
## measurement_id dragon_id date total_body_length_cm wingspan_cm
## <int> <chr> <chr> <dbl> <dbl>
## 1 1 D96 2012-10-10 1070. 1390.
## 2 2 D400 2003-07-03 333. 635.
## 3 3 D316 2017-09-08 867. 1052.
## 4 5 D484 2016-12-04 1032. 1721.
## 5 6 D149 2012-02-13 920. 1534.
## 6 7 D285 2016-03-23 305. 699.
## 7 8 D256 2013-09-07 359. 652.
## 8 9 D283 2007-06-21 1698. 1387.
## 9 10 D213 2001-12-12 354. 671.
## 10 11 D485 2002-10-04 957. 1780.
## # ℹ 254 more rows
## # ℹ 7 more variables: tail_length_cm <dbl>, tarsus_length_cm <dbl>,
## # claw_length_cm <dbl>, sex <chr>, age_class <chr>, species <chr>,
## # update_timestamp <chr>13.2.12 Conditional value assignment
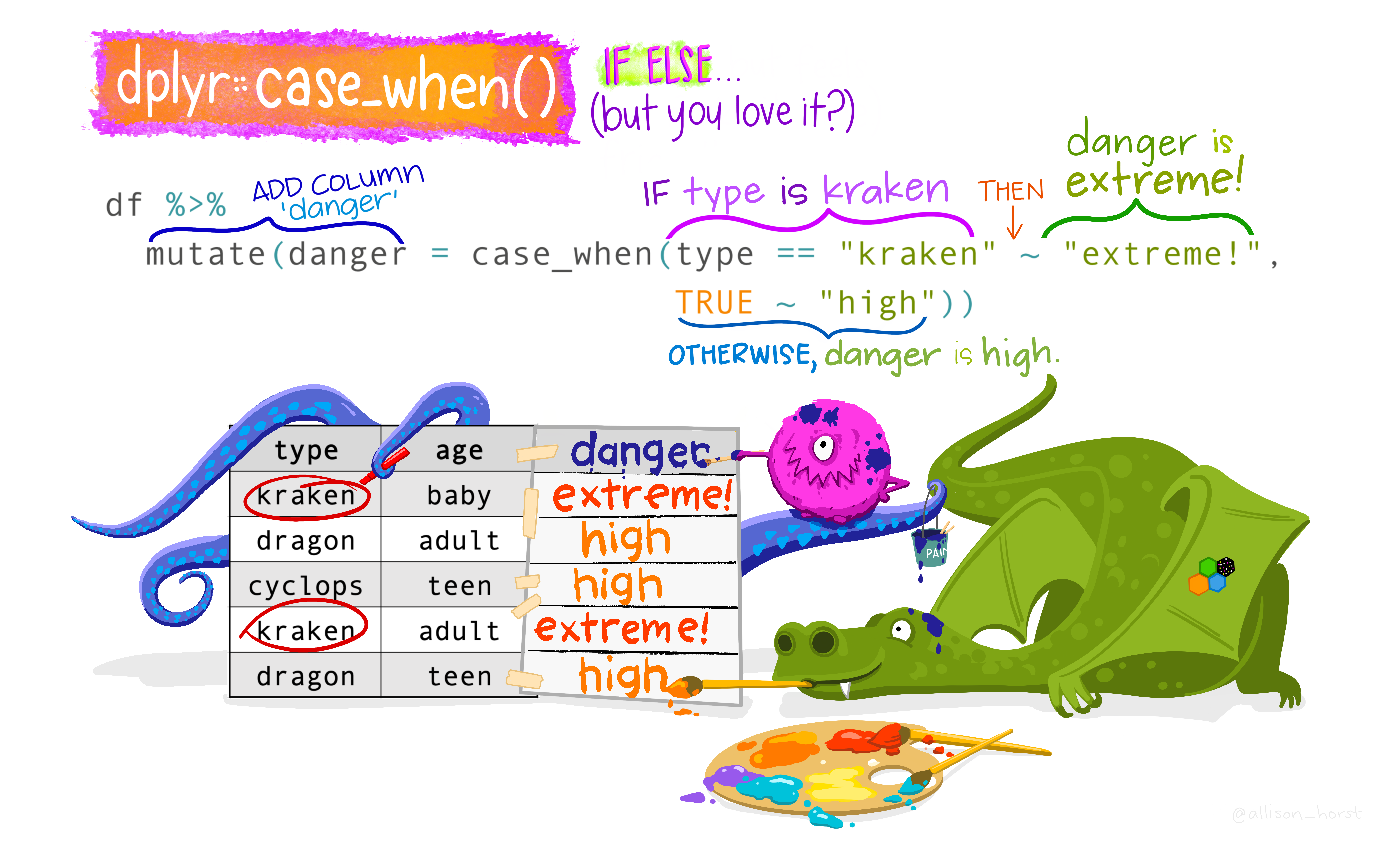
Figure 13.3: Artwork by Allison Horst
The last function we are going to look at is case_when. This function allows
to do conditional value assignment by vectorizing multiple statements of the
kind: if … else …. Let’s look at an example to understand what this
means. The dragons table includes information on sex and on age class. Say that
we wanted to create a composite variable combining sex and age class, with
possible categories “adult female”, “adult male”, “juvenile female”, etc. We can
gather the information from the two existing column and create a new column
assigning the appropriate category to each individual using case_when:
morphometrics %>%
as_tibble() %>%
left_join(dragons, by = "dragon_id") %>%
mutate(sex_age_combo = case_when(
sex == "F" & age_class == "Adult" ~ "Adult Female",
sex == "M" & age_class == "Adult" ~ "Adult Male",
sex == "F" & age_class == "Subadult" ~ "Subadult Female",
sex == "M" & age_class == "Subadult" ~ "Subadult Male",
sex == "F" & age_class == "Juvenile" ~ "Juvenile Female",
sex == "M" & age_class == "Juvenile" ~ "Juvenile Female",
is.na(sex) & age_class == "Adult" ~ "Adult Unknown",
is.na(sex) & age_class == "Subadult" ~ "Subadult Unknown",
is.na(sex) & age_class == "Juvenile" ~ "Juvenile Unknown"
)) %>%
select(dragon_id, sex, age_class, sex_age_combo)## # A tibble: 327 × 4
## dragon_id sex age_class sex_age_combo
## <chr> <chr> <chr> <chr>
## 1 D96 F Adult Adult Female
## 2 D400 <NA> Juvenile Juvenile Unknown
## 3 D316 M Subadult Subadult Male
## 4 D317 F Adult Adult Female
## 5 D484 M Adult Adult Male
## 6 D149 M Adult Adult Male
## 7 D285 <NA> Juvenile Juvenile Unknown
## 8 D256 <NA> Juvenile Juvenile Unknown
## 9 D283 F Adult Adult Female
## 10 D213 <NA> Juvenile Juvenile Unknown
## # ℹ 317 more rowsLet’s break down the syntax I used in the case_when statement. Each entry has
the form logical condition ~ value to assign. Let’s isolate the left-hand
side of the first entry:
## [1] FALSE FALSE TRUE TRUE FALSE TRUEThere can be as many entries as we want, but they should fully cover the
logical domain of the statement. In other words, we need to account for every
possibility. If, say, we left out the male subadult combination, case_when is
going to automatically assign NA:
morphometrics %>%
as_tibble() %>%
left_join(dragons, by = "dragon_id") %>%
mutate(sex_age_combo = case_when(
sex == "F" & age_class == "Adult" ~ "Adult Female",
sex == "M" & age_class == "Adult" ~ "Adult Male",
sex == "F" & age_class == "Subadult" ~ "Subadult Female",
sex == "F" & age_class == "Juvenile" ~ "Juvenile Female",
sex == "M" & age_class == "Juvenile" ~ "Juvenile Female",
is.na(sex) & age_class == "Adult" ~ "Adult Unknown",
is.na(sex) & age_class == "Subadult" ~ "Subadult Unknown",
is.na(sex) & age_class == "Juvenile" ~ "Juvenile Unknown"
)) %>%
select(dragon_id, sex, age_class, sex_age_combo)## # A tibble: 327 × 4
## dragon_id sex age_class sex_age_combo
## <chr> <chr> <chr> <chr>
## 1 D96 F Adult Adult Female
## 2 D400 <NA> Juvenile Juvenile Unknown
## 3 D316 M Subadult <NA>
## 4 D317 F Adult Adult Female
## 5 D484 M Adult Adult Male
## 6 D149 M Adult Adult Male
## 7 D285 <NA> Juvenile Juvenile Unknown
## 8 D256 <NA> Juvenile Juvenile Unknown
## 9 D283 F Adult Adult Female
## 10 D213 <NA> Juvenile Juvenile Unknown
## # ℹ 317 more rowsOnce we’ve covered all of the possibilities except one, we can leave the last
one implicit by using TRUE on the left-hand side of the last entry of the
case_when statement. For example, if an individual does not fall in the first
six categories it means its sex is unknown. We could lump these all together in
one “Unknown” category:
morphometrics %>%
as_tibble() %>%
left_join(dragons, by = "dragon_id") %>%
mutate(sex_age_combo = case_when(
sex == "F" & age_class == "Adult" ~ "Adult Female",
sex == "M" & age_class == "Adult" ~ "Adult Male",
sex == "F" & age_class == "Subadult" ~ "Subadult Female",
sex == "M" & age_class == "Subadult" ~ "Subadult Male",
sex == "F" & age_class == "Juvenile" ~ "Juvenile Female",
sex == "M" & age_class == "Juvenile" ~ "Juvenile Female",
TRUE ~ "Unknown"
)) %>%
select(dragon_id, sex, age_class, sex_age_combo)## # A tibble: 327 × 4
## dragon_id sex age_class sex_age_combo
## <chr> <chr> <chr> <chr>
## 1 D96 F Adult Adult Female
## 2 D400 <NA> Juvenile Unknown
## 3 D316 M Subadult Subadult Male
## 4 D317 F Adult Adult Female
## 5 D484 M Adult Adult Male
## 6 D149 M Adult Adult Male
## 7 D285 <NA> Juvenile Unknown
## 8 D256 <NA> Juvenile Unknown
## 9 D283 F Adult Adult Female
## 10 D213 <NA> Juvenile Unknown
## # ℹ 317 more rowsOr we could decide that if sex is unknown we just simply assign the value of
age_class:
morphometrics %>%
as_tibble() %>%
left_join(dragons, by = "dragon_id") %>%
mutate(sex_age_combo = case_when(
sex == "F" & age_class == "Adult" ~ "Adult Female",
sex == "M" & age_class == "Adult" ~ "Adult Male",
sex == "F" & age_class == "Subadult" ~ "Subadult Female",
sex == "M" & age_class == "Subadult" ~ "Subadult Male",
sex == "F" & age_class == "Juvenile" ~ "Juvenile Female",
sex == "M" & age_class == "Juvenile" ~ "Juvenile Female",
TRUE ~ age_class
)) %>%
select(dragon_id, sex, age_class, sex_age_combo)## # A tibble: 327 × 4
## dragon_id sex age_class sex_age_combo
## <chr> <chr> <chr> <chr>
## 1 D96 F Adult Adult Female
## 2 D400 <NA> Juvenile Juvenile
## 3 D316 M Subadult Subadult Male
## 4 D317 F Adult Adult Female
## 5 D484 M Adult Adult Male
## 6 D149 M Adult Adult Male
## 7 D285 <NA> Juvenile Juvenile
## 8 D256 <NA> Juvenile Juvenile
## 9 D283 F Adult Adult Female
## 10 D213 <NA> Juvenile Juvenile
## # ℹ 317 more rows13.3 Style
Tidyverse syntax helps to make code readable, but half of the deal with readability has to do with style. Hadley Whickham, the inventor of the tidyverse, has put together a handy style guide that lists things to pay attention to when writing code and best practices to adopt. Style is not so much about making things aesthetically pleasing (although it certainly does), but about making the code easy for your eyes to navigate through and understand. R already uses a type of font for which each character occupies the same amount of pixels, which makes things align nicely across rows. This makes it easy to recognize structure in the code at a glance, and this physical structure corresponds to logical structure so it helps you understand how the pieces fit together. But these qualities are no good if the user doesn’t put in the effort to take advantage of them. Here are my favorite style tips to make sure your code looks clean and reads easily:
- Always put spaces after your commas, on both sides of equal signs and other
operators (e.g., do
x <- 1, notx<-1); - Do not exceed 80 characters per line in your script (you can check how many characters you’ve used at the bottom-left corner of the script panel, and recent versions of RStudio also have a handy gray vertical line that marks the 80 character limit);
- Be consistent in your variable names: don’t use
CamelCasefor some andlowercase_with_underscoresfor others. Actually, stick tolowercase_with_underscores. - Start a new line after each pipe
%>%; - I like to start a new line even after each column listed in a
selectstatement, or even after each argument of a function (for sure whenever the code would overflow past 80 characters otherwise); - Tidyverse automatically indents your code when you go to a new line (e.g.,
after a pipe), but if you happen to mess with the alignment while you’re editing
the code, make sure to reindent it and realign it at the end (you don’t have to
do it manually, just highlight the chunk of code you want to reindent and go to
Code > Reindent lines in RStudio, or press
Ctrl+I).